Coding Best Practices
Ensuring a Reproducible workflow
A very important concept in statistical data analysis (and in programming and science in general) is the concept of a reproducible workflow. What this means is that, given your code, anyone (including your future self) should be able to reproduce your results exactly.
To be more specific, imagine this scenario:
- You are a data analyst at a company, and your manager tells you “hey, I really liked those graphs you made for your presentation last week, can you make these important changes and send it to me by lunchtime? I need to brief the C-Suite at 1pm”.
- You get yourself a coffee, sit down at your machine, fire up your RStudio, and open up the file that you worked on last week late into the night. You try to run your script again, and to your horror, you find numerous errors.
- You try to reconstruct your thought processes that night, as you click into this menu and that menu. “Did I click this checkbox or not when I imported the data?”. “Did I import
data_version1ordata_version2?”. “How come this function is giving me a different result now? Or is throwing an error now when it didn’t last week?” - If you are lucky you are able to retrace every click you did. If not, you’ll have to tell your boss that you can’t reproduce that graph, let alone make the changes. Regardless, there will be a lot of mental and emotional cost that was unnecessary.
Now imagine this other scenario:
- You fire up your RStudio, hit a button, and your code automagically produces a nicely formatted report with the graph you made last week. You go into your code, modify a few lines to make the changes your manager wants. You run your code again, get the new graph out, send it to your manager, and happily move on to your next task.
Consider this last scenario:
- Your manager asks you for some additional analysis on James’ project, but James had left the company last week. James had given you all his code, neatly organized.
- You plop James’ code into your RStudio, and hit “run”. It magically works!
- Or, maybe it doesn’t.
- Which world do you want to be in?
- By the way, James doesn’t even have to have left the company — he could be a colleague in a remote team, or on leave that week, or otherwise indisposed to do that analysis, and you have to pick up where he left off. These types of situations happen all the time.
- Isn’t it much better for everyone to have a workflow that works again and again?
What is a reproducible workflow
What a reproducible workflow means is:
- You should write every part of your data analysis pipeline into code.
- This includes setting your working directory.
- It is very handy to go to the point-and-click interface to set your working directory, since your file may reside deep in many other folders. For example, on Mac, I can click
Session->Set Working Directory->To Source File Location. - After you click this, RStudio will output a
setwd(...)command in the console. For example:setwd("~/stats-notes"). Take this line, and copy it into the preamble of your R script, so that when you next open the file, you know which is your working directory and you can just set it by running that line.- Note: R Markdown has a different way of handling working directories. When you knit a Rmd file, it automatically assumes the working directory of the source file.
- It is very handy to go to the point-and-click interface to set your working directory, since your file may reside deep in many other folders. For example, on Mac, I can click
- This also includes reading in your data.
- Do NOT use the Import Dataset from the dropdown menu.
- DO Use
read.csv(...)orread.xlsx(...)or equivalent commands to read in your datasets.
- This also includes loading libraries and packages.
- Do NOT use the
Packagespane of RStudio to click and load the packages you require for your analysis. - DO use
library(...)to load in packages.
- Do NOT use the
- This includes setting your working directory.
For more advanced analysts (including graduate-level researchers preparing publication-quality analyses), you may also want to consider:
- Putting all your data processing / data cleaning steps into code. So your code should take in the raw datafile (as raw as possible) and manipulating it only in R. (NEVER EXCEL).
- If data processing takes too long to run every time, you could output a “cleaned” datafile and just read that in everytime you do analysis. But you should still save the data processing code just in case you need to modify it or check what you did.
- This can be saved either in another R script or in a chunk in an R Markdown file with
eval=Fso that it won’t run every single time.
- Putting any exclusion criteria into code. This also allows you to modify your exclusion criteria if you needed to.
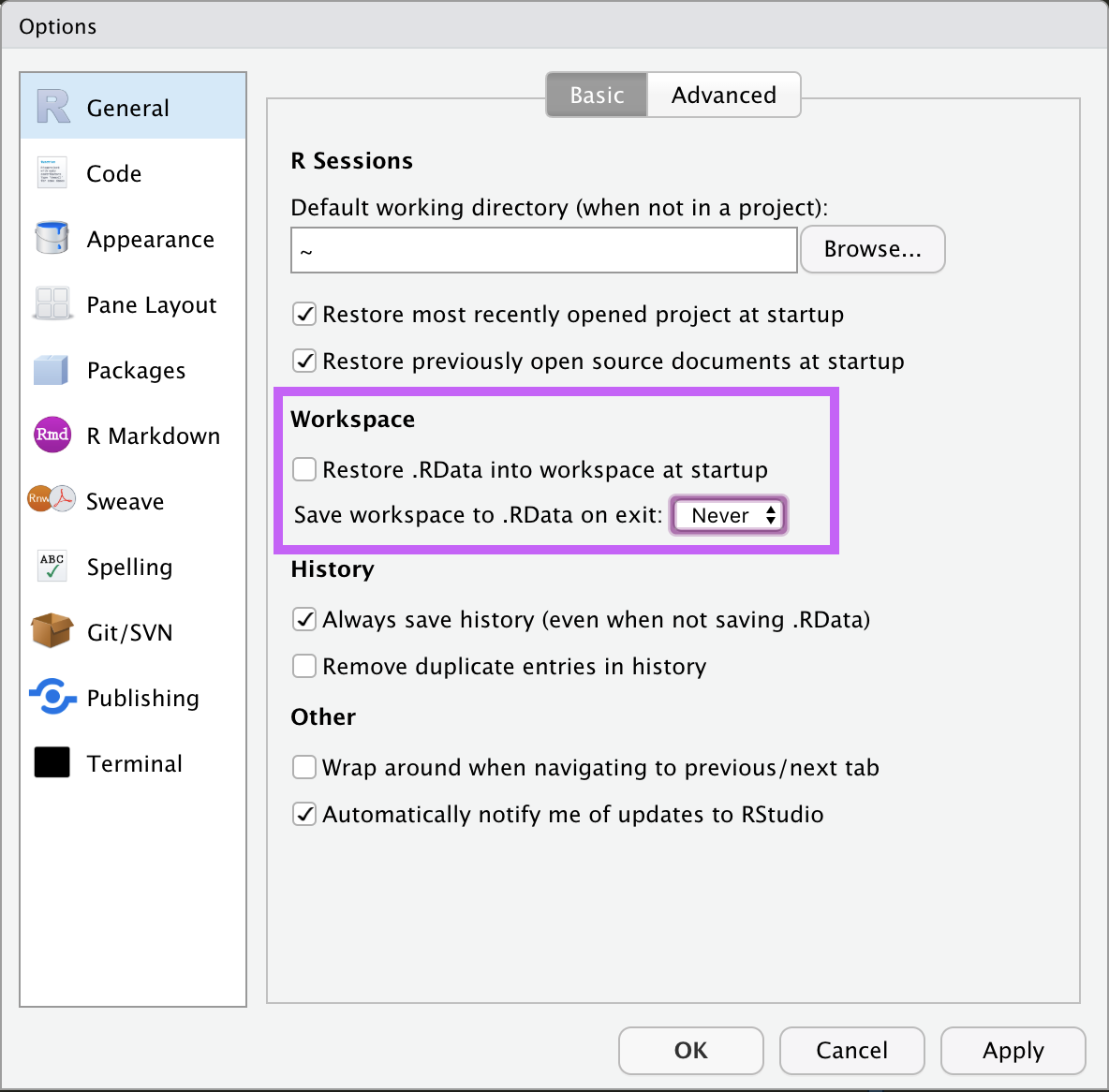
DO NOT SAVE .RDATA!
A pre-requisite for a reproducible workflow is that everytime you fire up RStudio, it should be a clean session, with no data loaded and no libraries loaded.
This means that you should NOT save or reload your session. If RStudio ever prompts you to save your session (to a .RData file) upon exit, do not accept.
You can customize RStudio to never do this by going into Preferences (see screenshot below for Mac OSX) and making sure:
- the
Restore .RData into workspace at startupis unselected. - the
Save workspace to .RData on exitdropdown menu is set toNever.
Building habits to ensure a reproducible workflow takes time, but you will thank yourself later for this. It’s never too early to start, and in fact, the earlier the better, so you do not have to unlearn bad habits. It’s just like, for programming, learning programming conventions: why do people name their variables like this or structure their code like that? After you become much more proficient in programming, you’ll see why it helps to make your code so much more efficient. This is the same with reproducibility.
Other Coding Best Practices
(This will be a growing list that I will add to.)
- Try not to re-use variable names or data frame names.
- I highly recommend NOT doing something like:
d1$score = d1$score * 100.- Why? Well, if you accidentally run that line of code again, your
scorewill be 100x higher than you wanted it to be.
- Why? Well, if you accidentally run that line of code again, your
- I highly recommend NOT doing something like:
- Try having longer and more informative names.
- Do NOT write something like:
mean = mean(d1$score).- Why? Doing this will overwrite
mean()(the function that calculates the average) in the namespace withmean(the variable that you just created). This means (haha) that you will not be able to calculate any more means because typingmeanwould then reference the newly-created variable, not the function you had in mind. - Instead, try to be more informative
meanScore = mean(d1$score). If ever in doubt, just type the variable name into the console to see if it’s already used. If it is, it’ll print out a function or some data. If not, it’ll sayError: object 'x' not found.
- Why? Doing this will overwrite
- Do NOT write something like:
- Do not use very common letters like x, y, i, j. Especially as counter variables.
- For example, what’s wrong with the following code?
x = mean(d1$score)...for(x in 1:10) {print(d1$score[x])}
- Then later when you want to print out the mean score, you’ll find that
xis 10. And not what you expected. - I personally recommend using longer variable names, even for counter variables, and again, never re-using them.
- For example, what’s wrong with the following code?
Analysis Best Practices
(This will be a growing list that I will add to.)